Ongoing Projects
Cell Biology of the Last Eukaryote Common Ancestor (LECA)
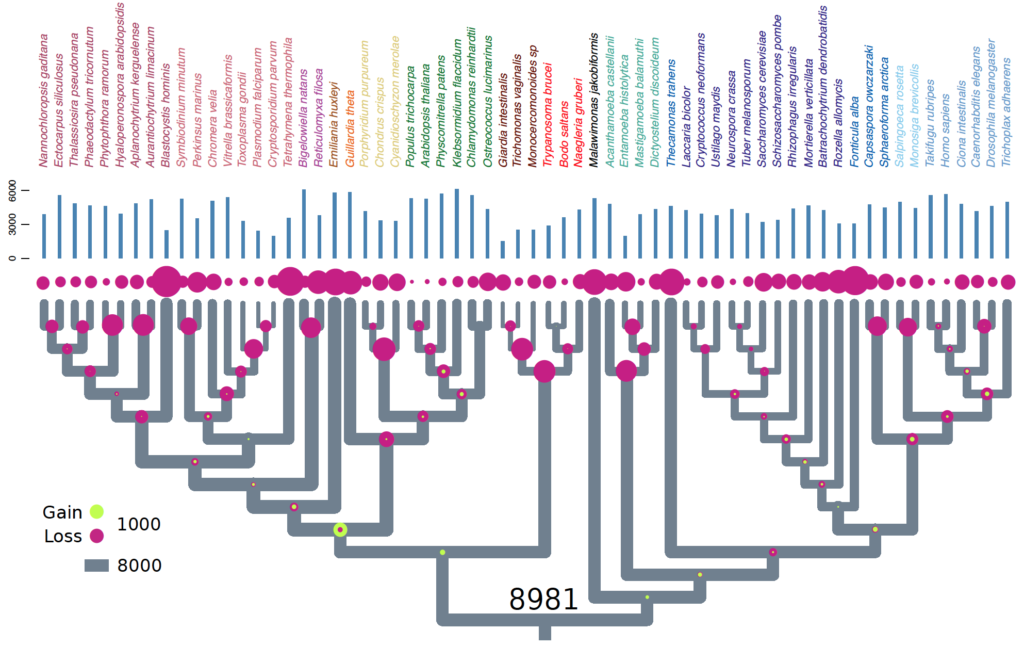
Eukaryotes are complex cells containing membrane bound organelles. The origin of eukaryotic cell complexity is shrouded in mystery due to the paucity of extant intermediates. The last eukaryote common ancestor (LECA) existed ~2 billion years ago. LECA was not a simple organism. It was a ‘ready-made’ eukaryote with all the bells and whistles. It had a nucleus, mitochondria, flagella, complex endomembrane and cytoskeletal systems. It was capable of performing such feats as mitosis, meiosis + cell fusion (sex). And finally it was likely bacteriovorous and had a complex lifecycle with multiple stages. In short, LECA resembled a modern-day free-living heterotrophic flagellated protist. Since nearly all molecular cell biological data originates from work on animal, plant, and fungal models, our understanding of LECA, and heterotrophic flagellates in general is extremely lacking.
Single cell genomics of heterotrophic flagellates
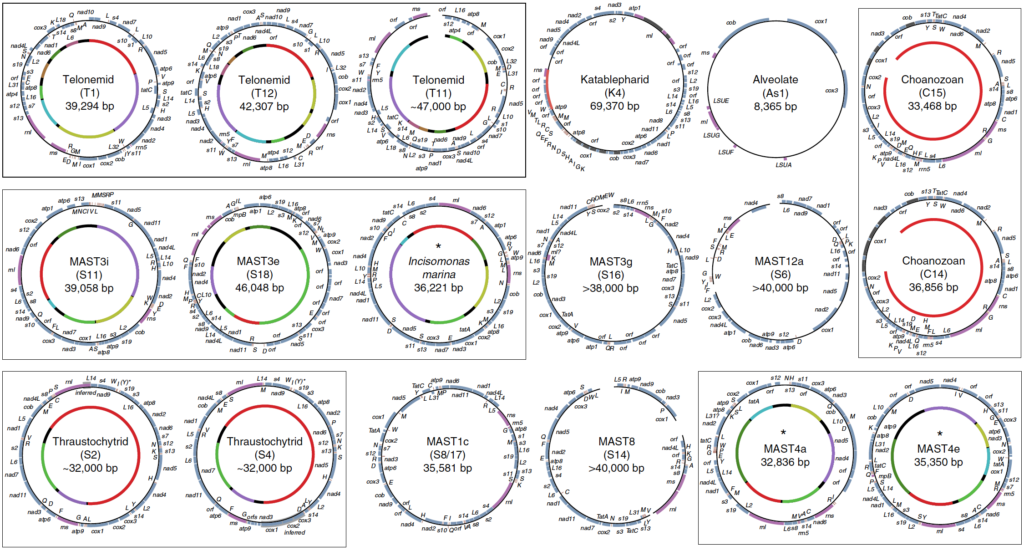
In order to gain insight into the biology of heterotrophic flagellates, more genomic and transcriptomic information from diverse lineages is needed. Heterotrophic flagellates are found in every major linage of eukaryotes, and in fact, every major lineage of eukaryotes evolved from a heterotrophic flagellate, meaning that major lineages that are not heterotrophic flagellates have sister lineages that are. Thus, in order to reconstruct more accurately the characteristics of LECA, the most basally diverging branches from each lineage must be sampled. Problematically some of these lineages have not yet been discovered and may have even gone extinct. Recent advances in single-cell genomics have vastly improved our ability to sample rare cells in natural environments leading to important discoveries in cell evolution of particular eukaryotic groups. Previous studies have focused primarily on marine ecosystems. Our current work is investigating the diversity and evolution of heterotrophic flagellates in freshwater and desert soil environments.
Evolution and diversification of mitochondrial form and function
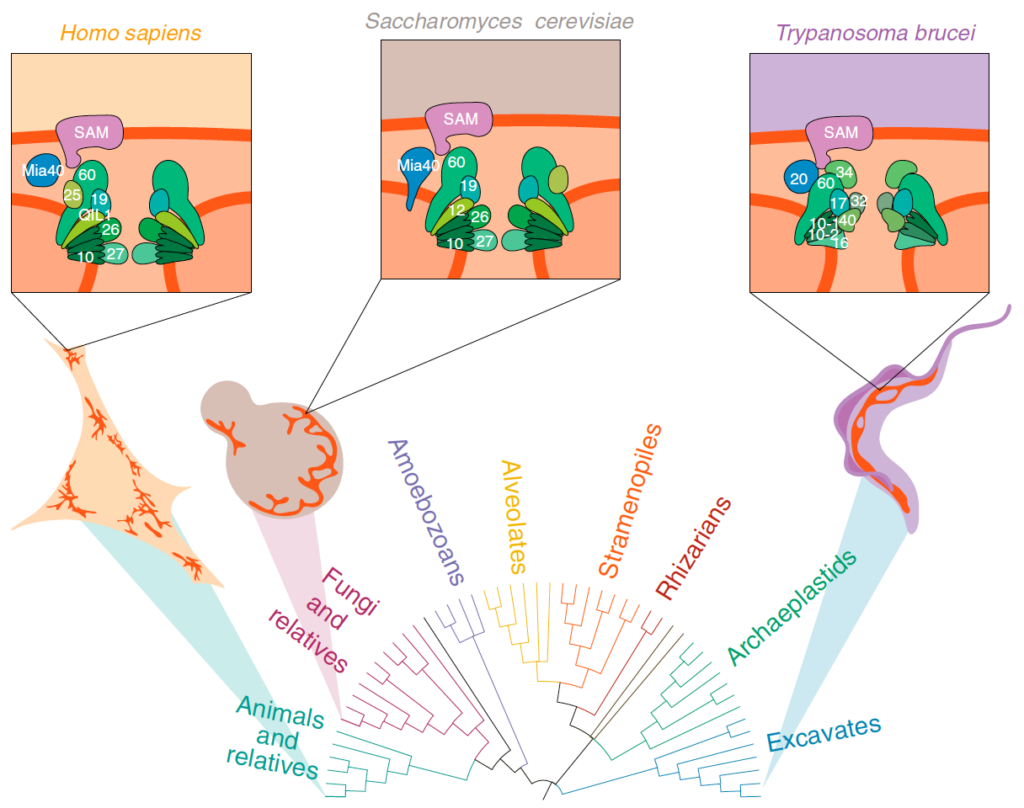
Mitochondria are known for housing the electron transport chain and being the site of ATP synthesis in eukaryotic cells. However, their cell biology is much more complicated than just their biochemical prowess. Mitochondria in many lineages constantly move, fuse, and divide; however, whether these behaviours arose in LECA or later convergently, is unclear. Mitochondrial cristae membranes – the sites of electron transport and ATP synthesis – have diverse morphologies in different cell types and lineages. How and why these varied cristae morphologies evolved is unclear – we do not understand the functional implications of this variation. Finally, although mitochondria contain their own genomes, most of their protein complement is encoded by the nuclear genome and is imported and assembled into mitochondrial compartments by dedicated translocation machinery. Most of this machinery can be traced back to LECA, but translocase sequences vary wildly between lineages. Whether this variation is due to selective pressures or drift is unknown. To gain insight into the evolution of mitochondrial dynamics, cristae morphology, and protein import, we use Saccharomyces cerevisiae as a tool for heterologous expression of mitochondrial proteins from diverse eukaryotes.